Alternatively, you can use IGMH (independent gradient model based on Hirshfeld partition) instead, the graphical effect is similar to RDG, but you can manually define fragments and only portray interfragment interactions, see Section 4.20.11 of Multiwfn manual for example.
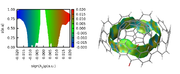
]]>I want to use the NCI method and exactly RDG (first subfunction of function 20 in the latest version) to visualize the non-covalent interactions in a dimer, exactly like this picture
I need to note that I don't want to use "promolecular density method". I just want to use the RDG method.
the problem is that when I try to use the RDG method, there is no option to define two fragments to visualize only the interactions between these parts and not in the whole system.
please help to solve this problem.
Thank you