New content has been added to Section 4.17.2 of Multiwfn manual:
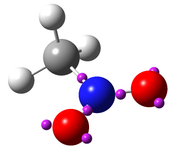
The attractors located by Multiwfn can be visualized via third-part softwares. To visualize them in VMD (http://www.ks.uiuc.edu/Research/vmd/), you should choose "-4 Export attractors as pdb/pqr/txt/gjf file" and select corresponding option to export all attractors as .pdb or .pqr file, which can be loaded and plotted VMD (PS: In .pqr file, the atomic charge column corresponds to function value at the attractor). The atoms and attractors can also be exported as .gjf using corresponding suboption, then you can use GaussView to easily visualize attractors. After loading the .gjf file into GaussView, it is suggested to choose "File" - "Preference" - "View" - "Display Format" - "Molecule", and then set low layer as "Tube" style. It is also suggested to disable showing labels in GaussView. The following map shows ELF attractors of CH3NO2 displayed in this way:
]]>
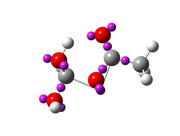
I followed your instructions, but the results was disappointing, i don't know where the problem come from ? Why the Multiwfn GUI show a good results, but Gaussview can't ?
I hope in the futur that Multiwfn support Gaussview.
My warmest thanks.
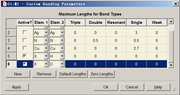
]]>If you do not hope gview to automatically add bonds between dummy atoms and real atoms, you can enter "Tools" - "Custom Bonding parameter", and add new terms defining the bonding threshold between X and existing elements, then click "Apply" to take effect, the unwanted bonds will be cleaned. Note that this function is only available since GaussView 6.0.
]]>I found this article about the same question (http://bbs.keinsci.com/thread-16326-1-1.html), I follow your instructions, but i got unsatisfying results.
]]>Multiwfn is great and a wonderful program, and I just performed my first analysis using the program (ELF analysis) by following the tutoriel of Mr. Sobereva, but i don't like like the visualisation using VMD, and i wonder if i thers is a trick that make to visualize the results using Gaussview? I tried, but i didn't succeed, the bonds between carbon atoms and the attractors was too short.
Many thanks in advance
Maximos